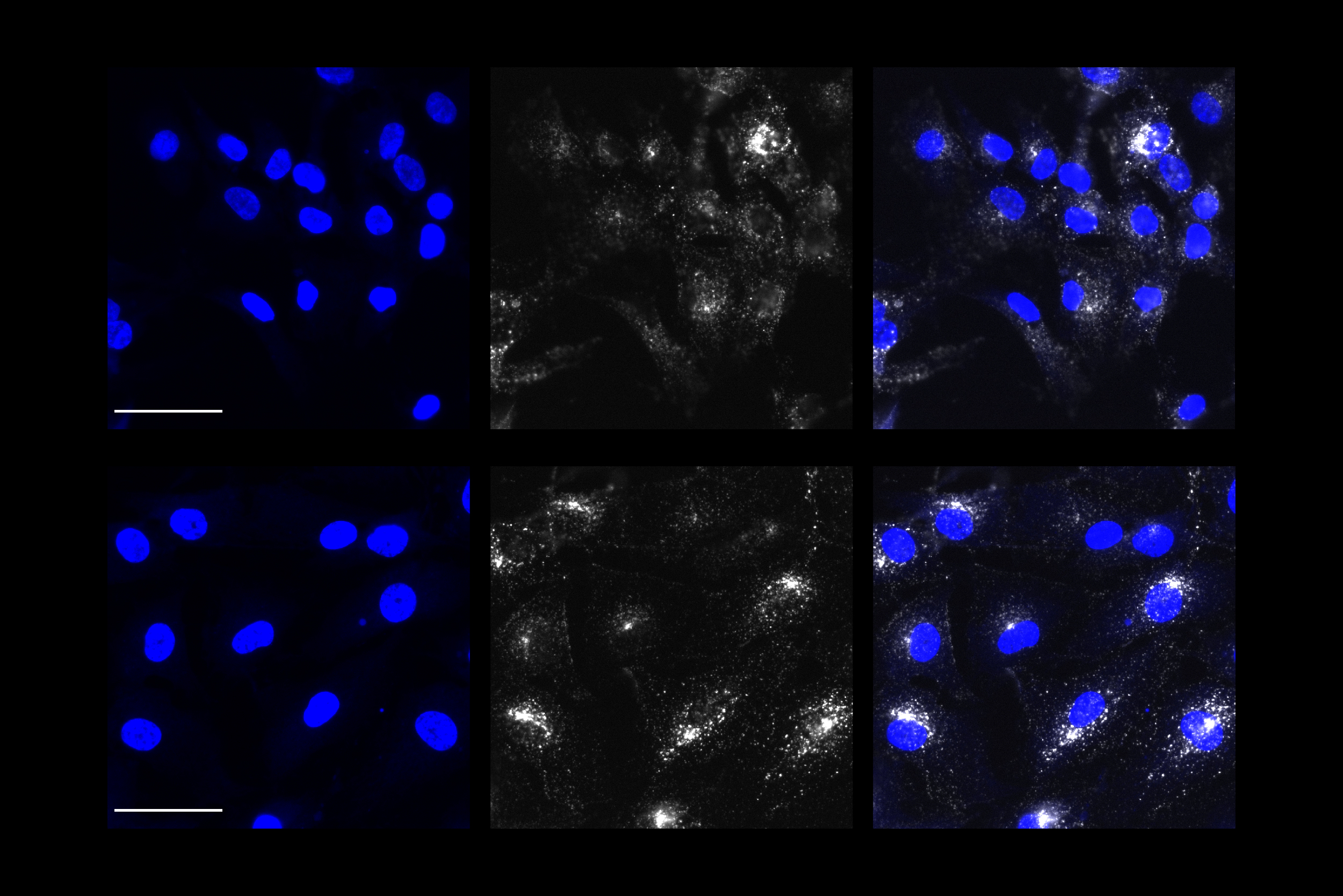
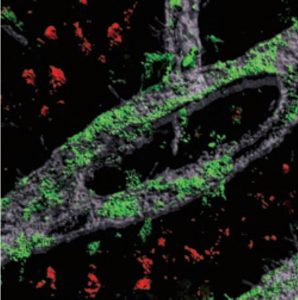
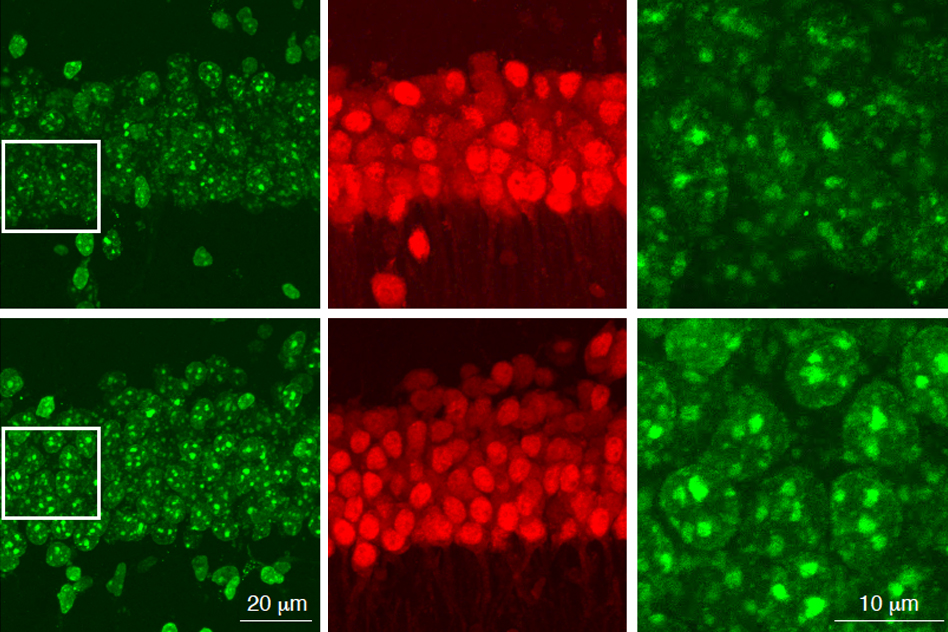
The findings may help explain why some people who lead enriching lives are less prone to Alzheimer’s and age-related dementia
Many people develop Alzheimer’s or other forms of dementia as they get older. However, others remain sharp well into old age, even if their brains show underlying signs of neurodegeneration.
Among these cognitively resilient people, researchers have identified education level and amount of time spent on intellectually stimulating activities as factors that help prevent dementia. A new study by MIT researchers shows that this kind of enrichment appears to activate a gene family called MEF2, which controls a genetic program in the brain that promotes resistance to cognitive decline.
The researchers observed this link between MEF2 and cognitive resilience in both humans and mice. The findings suggest that enhancing the activity of MEF2 or its targets might protect against age-related dementia
“It’s increasingly understood that there are resilience factors that can protect the function of the brain,” says Li-Huei Tsai, director of MIT’s Picower Institute for Learning and Memory. “Understanding this resilience mechanism could be helpful when we think about therapeutic interventions or prevention of cognitive decline and neurodegeneration-associated dementia.”
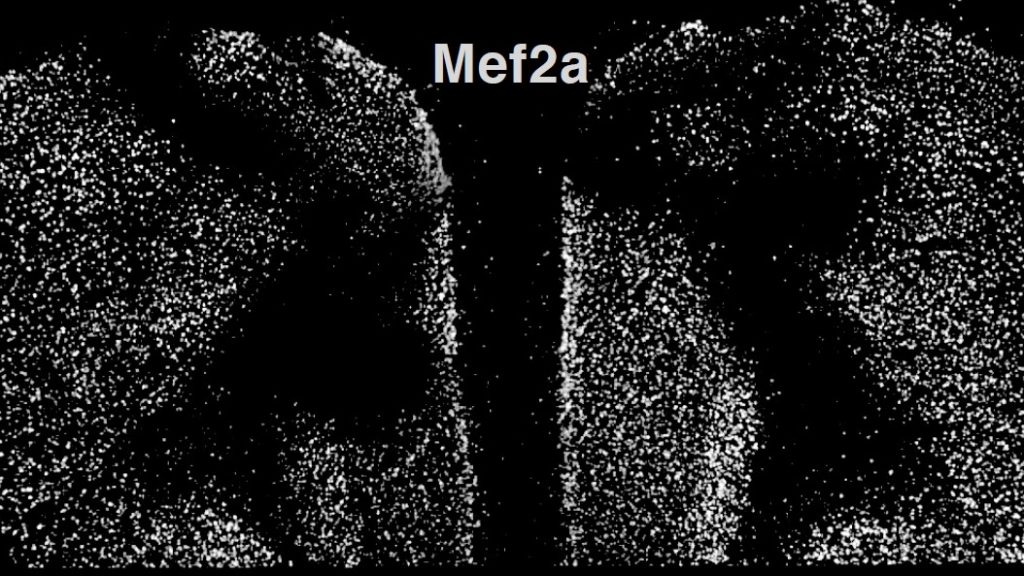
Above: When researchers knocked down a Mef2 gene in the frontal cortex of mice (blank areas), it prevented the mice from attaining a cognitive benefit of being raised in an enriched environment.
Tsai, a founding director of MIT’s Aging Brain Initiative, is the senior author of the study, which appears today in Science Translational Medicine. The lead authors are recent MIT PhD recipient Scarlett Barker and MIT postdoctoral fellow and Boston Children’s Hospital physician Ravikiran (Ravi) Raju.
Protective effects
A large body of research suggests that environmental stimulation offers some protection against the effects of neurodegeneration. Studies have linked education level, type of job, number of languages spoken, and amount of time spent on activities such as reading and doing crossword puzzles to higher degrees of cognitive resilience.
The MIT team set out to try to figure how these environmental factors affect the brain at the neuronal level. They looked at human datasets and mouse models in parallel, and both tracks converged on MEF2 as a critical player.
MEF2 is a transcription factor that was originally identified as a factor important for cardiac muscle development, but later was discovered to play a role in neuron function and neurodevelopment. In two human datasets comprising slightly more than 1,000 people all together, the MIT team found that cognitive resilience was highly correlated with expression of MEF2 and many of the genes that it regulates.
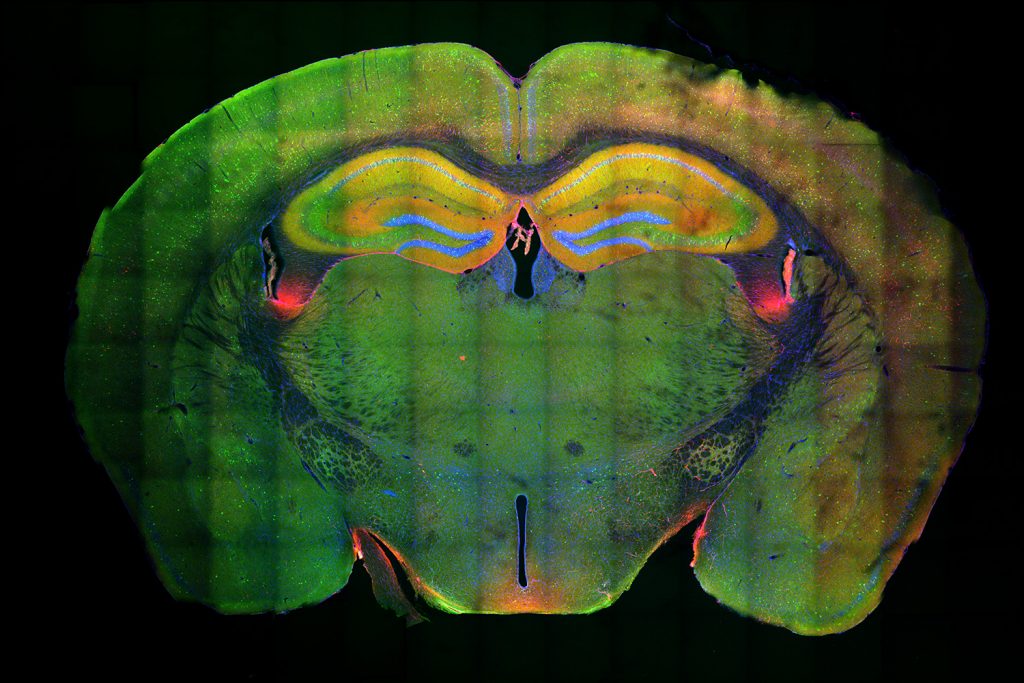
Many of those genes encode ion channels, which control a neuron’s excitability, or how easily it fires an electrical impulse. The researchers also found, from a single-cell RNA-sequencing study of human brain cells, that MEF2 appears to be most active in a subpopulation of excitatory neurons in the prefrontal cortex of resilient individuals.
To study cognitive resilience in mice, the researchers compared mice who were raised in cages with no toys, and mice placed in a more stimulating environment with a running wheel and toys that were swapped out every few days. As they found in the human study, MEF2 was more active in the brains of the mice exposed to the enriched environment. These mice also performed better in learning and memory tasks.
When the researchers knocked out the gene for MEF2 in the frontal cortex, this blocked the mice’s ability to benefit from being raised in the enriched environment, and their neurons became abnormally excitable.
“This was particularly exciting as it suggested that MEF2 plays a role in determining overall cognitive potential in response to variables in the environment,” Raju says.
The researchers then explored whether MEF2 could reverse some of the symptoms of cognitive impairment in a mouse model that expresses a version of the tau protein that can form tangles in the brain and is linked with dementia. If these mice were engineered to overexpress MEF2 at a young age, they did not show the usual cognitive impairments produced by the tau protein later in life. In these mice, neurons overexpressing MEF2 were less excitable.
“A lot of human studies and mouse model studies of neurodegeneration have shown that the neurons become hyperexcitable in early stages of disease progression,” Raju says. “When we overexpressed MEF2 in a mouse model of neurodegeneration, we saw that it was able to prevent this hyperexcitability, which might explain why they performed cognitively better than control mice.”
Enhancing resilience
The findings suggest that enhancing MEF2 activity could help to protect against dementia; however, because MEF2 also affects other types of cells and cellular processes, more study is needed to make sure that activating it wouldn’t have adverse side effects, the researchers say.
The MIT team now hopes to further investigate how MEF2 becomes activated by exposure to an enriching environment. They also plan to examine some of the effects of the other genes that MEF2 controls, beyond the ion channels they explored in this study. Such studies could help to reveal additional targets for drug treatments.
“You could potentially imagine a more targeted therapy by identifying a subset or a class of effectors that is critically important for inducing resilience and neuroprotection,” Raju says.
The research was funded by the Glenn Center for Biology of Aging Research, the National Institute of Aging, the Cure Alzheimer’s Fund, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
-From MIT News